 |
Affected male |
 |
Unffected male |
 |
Affected female |
 |
Unaffected female |
Pedigree file contains information about family relationships, gender (=sex) and genetic data (disease and marker phenotypes).
Pre-makeped format contains following columns, separated by space and/or tab characters:
Column 1: Pedigree identifier { The identifier can be a number
Column 2: Individual's ID { or a character string
Column 3: The individual's father { If the person is a founder, just put a
Column 4: The individual's mother { 0 in each column.
Column 5: Sex { 1 = Male, 2 = Female, Unknown sex is not permitted
Column 6+: Genetic data { Disease and marker phenotypes
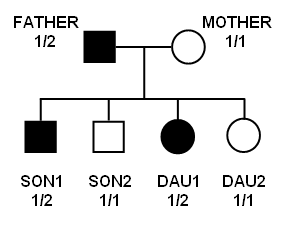
Let's assume following pedigree structure (with discrete trait / qualitative phenotype, one liability class) with father, mother, two sons (son1 and son2) and two daughters (dau1 and dau2) with one marker phenotype:
| Pedigree | Pedigree symbols | ||||||||
 |
|
Pedigree structure coded to text file (pre-makeped):
ped1 father 0 0 1 2 1 2
ped1 mother 0 0 2 1 1 1
ped1 son1 father mother 1 2 1 2
ped1 son2 father mother 1 1 1 1
ped1 dau1 father mother 2 2 1 2
ped1 dau2 father mother 2 1 1 1
Father's and mother's parents are unknown and their id is set to zero (=0) and after sex column is affection status column (disease) where number:
If disease locus has more than one liability class, then liability class column is right after disease. After disease locus column(s) comes marker phenotypes.
using numbers (recommended):
1 1 0 0 1 2 1 2
1 2 0 0 2 1 1 1
1 3 1 2 1 2 1 2
1 4 1 2 1 1 1 1
1 5 1 2 2 2 1 2
1 6 1 2 2 1 1 1
NOTE! Number of spaces between columns does not matter, but if it's formatted as above, it's easy to read. If some individual is not genotyped in some marker, you must enter 0 for each allele (e.g. phenotype = 0 0).
Last line of the file must be empty line!
::: AUTOGSCAN uses pre-makeped LINKAGE format! :::
Post-makeped format comes after above pedigree file is processed with makeped program, which is readable format for LINKAGE (FASTLINK) package and others. See makeped how-to for more info.
LINKAGE format contains following columns:
Column 1: Pedigree number
Column 2: Individual ID number
Column 3: ID of father
Column 4: ID of mother
Column 5: First offspring ID
Column 6: Next paternal sibling ID
Column 7: Next maternal sibling ID
Column 8: Sex
Column 9: Proband status (1=proband, higher numbers indicate doubled individuals formed
in breaking loops. All other individuals have a 0 in this field.)
Column 10+: Disease and marker phenotypes (as in the original pedigree file)
Here is above pedigree after it's processed with makeped:
1 1 0 0 3 0 0 1 1 2 1 2 Ped: ped1 Per: father
1 2 0 0 3 0 0 2 0 1 1 1 Ped: ped1 Per: mother
1 3 1 2 0 4 4 1 0 2 1 2 Ped: ped1 Per: son1
1 4 1 2 0 5 5 1 0 1 1 1 Ped: ped1 Per: son2
1 5 1 2 0 6 6 2 0 2 1 2 Ped: ped1 Per: dau1
1 6 1 2 0 0 0 2 0 1 1 1 Ped: ped1 Per: dau2
Makeped recoded pedigree and individual identifiers to numbers and added extra columns. Original pedigree and individual ID's are end of the each line.
It's recommended to have each chromosome in separate file and name files according to chromosome as chr1.raw, chr2.raw,...,chr22.raw.
Handbook of Human Genetic Linkage, Joseph D. Terwilliger and Jurg Ott. Johns Hopkins University Press, Baltimore (1994)